EcoScope researchers break new ground in AI-automated open science species distribution modelling
A team of EU-funded EcoScope project scientists published a paper detailing “An open science automatic workflow for multi-model species distribution estimation”, accessible online since March 7, 2024 in the International Journal of Data Science and Analytics. The research was published by EcoScope researchers Gianpaolo Coro, Lorenzo Sana and Pasquale Bove, of Italy’s National Research Council’s Faedo Institute for Information Science and Technology (ISTI-CNR).
“The unsustainable exploitation of ocean resources – with overfishing, chemical and physical pollution, and heavy maritime traffic – threatens oceans, seas, and coasts. Climate change further exacerbates this problem. Digital technologies are crucial tools to understand how to manage the effects of this pressure and potentially help mitigate it,” the researches commented. “Our workflow belongs to big data processing methodologies.”
Coro, Sana and Bove’s study presents a fully-automatic workflow for estimating species’ distributions through statistical and machine learning models, integrating four ecological niche models (ENMs) with complementary approaches: Artificial Neural Networks, Maximum Entropy, Support Vector Machines, and AquaMaps.
The workflow developed combines the models within one ensemble model, and automatically estimates the optimal model parameterisations and decision thresholds to distinguish between suitable and unsuitable habitat locations. Additionally, several ensemble models are combined to produce a mapped biodiversity index, reflecting species richness, the researchers noted.
Full automatisation makes the workflow particularly useful in automatically discovering mutual relations between the different forces stressing an ecosystem, which is also “a crucial focus” in designing Digital Twins of the Ocean (DTOs), the team said, adding that they intend to propose the workflow’s use in DTOs as well.
In the paper, the team focused specifically on a case study predicting the spread of the invasive fish species Siganus rivulatus in the Mediterranean and its future overlap with the native fish species Sarpa salpa under various climate change scenarios. The projections indicated that climate change will likely facilitate the further invasion of the Mediterranean basin by S. rivulatus and increase its overlap with S. salpa, increasing its risk of habitat loss and damage to fisheries, especially in the scenario of higher greenhouse gas emissions, the researchers concluded.
“Our workflow is general enough to process the data of other areas, species, and scenarios than those presented in the case studies. Moreover, it can integrate the outputs of additional ENMs,” they noted.
The researchers also assessed the workflow’s stability and sensitivity, and demonstrated its effectiveness by producing a Mediterranean biodiversity index, including models of 1508 European species.
Explaining the need for the automated workflow they developed, the team noted that Integrated Environment Assessment systems and ecosystem models have long implementation times, are susceptible to data-model interoperability issues, and require a range of heterogenous competencies.
Such models – which are a key tool for studying links between anthropogenic and natural climatic pressures on marine ecosystems and learning how to manage the effects of unsustainable exploitation – would therefore benefit from simplification, automatisation, and enhanced integrability of the underlying models, they said.
“Artificial intelligence can help overcome several limitations by speeding up the modelling of crucial functional parts,” they added, specifically, for example, in “estimating the environmental conditions fostering a species’ persistence and proliferation in an area (the species’ ecological niche) and, consequently, its geographical distribution.”
Importantly, the automated workflow’s software is open-source, Open Science compliant, and available as a web processing service-standardised cloud computing service, which enhances its efficiency, integrability, cross-domain reusability, and experimental reproduction and repetition, the researchers added.
“Open Science compliance requires the models to be available under recognised standards of interoperability and integrability and the published results to be repeatable and reproducible (after changing some model-parameter values),” they explained. Such features are crucial for guaranteeing the transparency of the results for decision-making authorities and promoting the consideration of the results in policy making, the researchers emphasised.
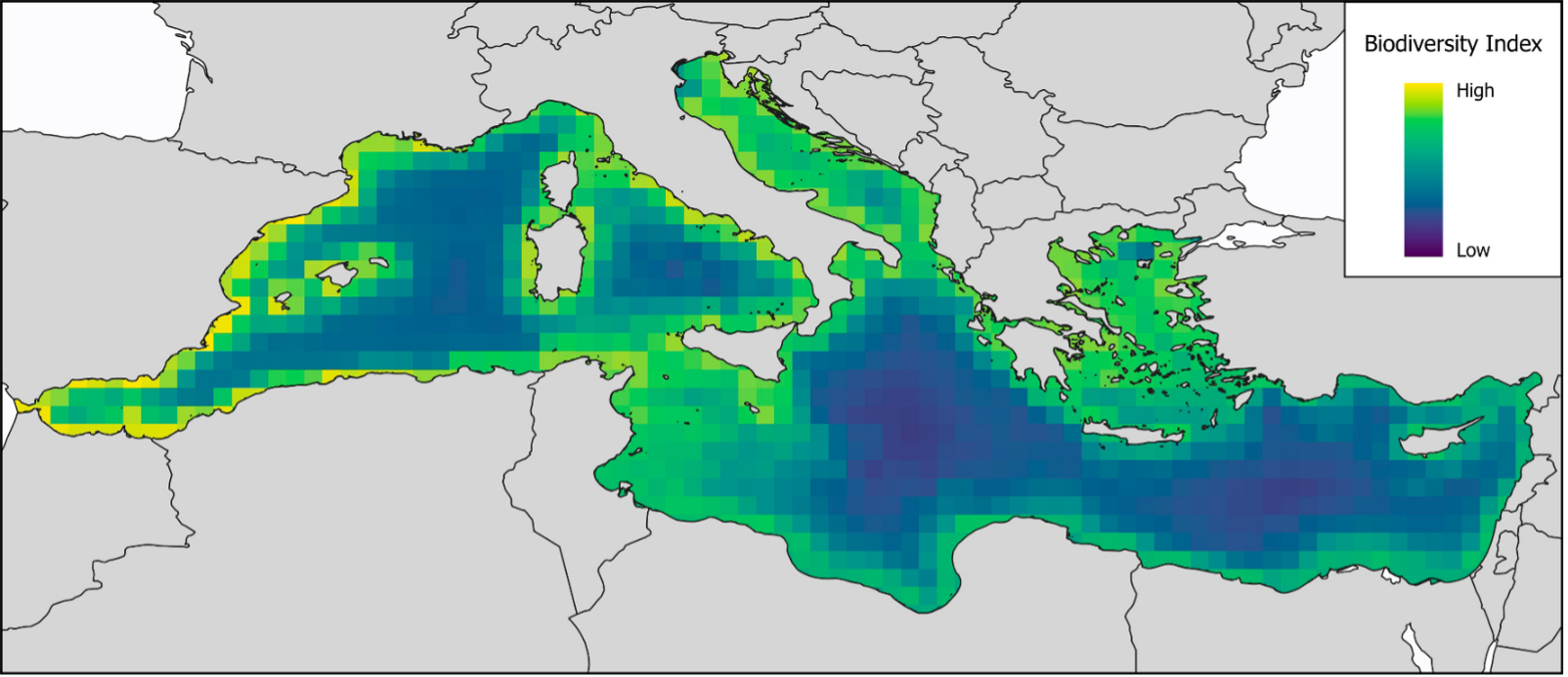
Biodiversity index (species richness) at half-degree resolution, produced by our workflow after processing 1508 Mediterranean species data
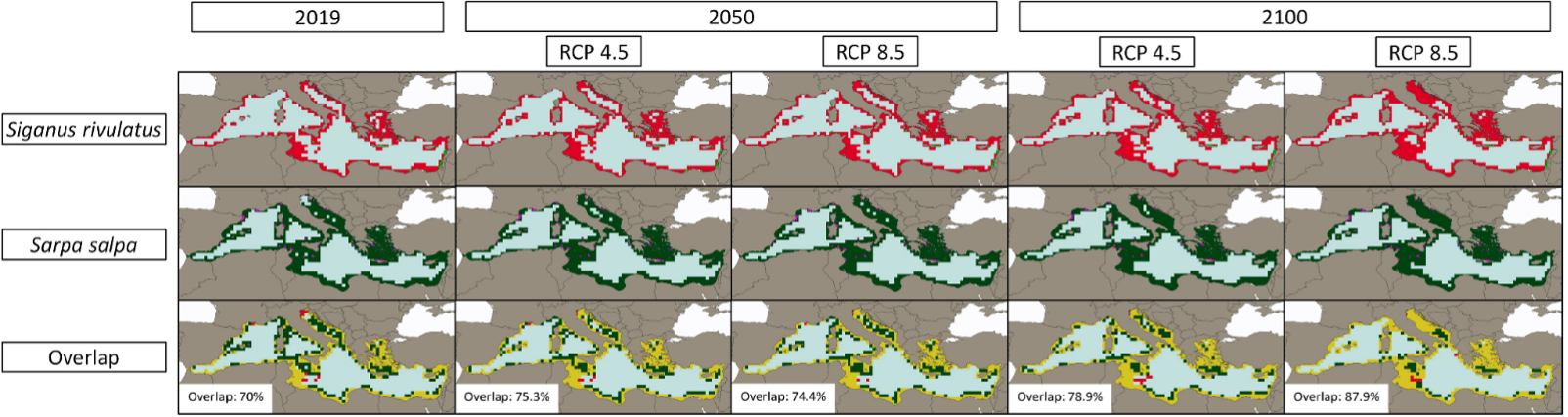
Per cent overlap between the estimated distributions of Siganus rivulatus and Sarpa salpa in 2019, 2050 and 2100. Future projections are reported for medium (RCP4.5) and high (RCP8.5) greenhouse gas emission scenarios. Small green dots report the S. rivulatus observations from OBIS, and purple dots those of S. salpa.





